Metadata
aliases: []
shorthands: {}
created: 2022-03-01 12:01:52
modified: 2022-07-11 13:29:30
Let's say we have a triangle mesh, stored using the separate arrays convention and we also have a set of points in space close to this surface and we want to evaluate the density of points on the mesh surface.
For this, we take every single point in the point set and project them orthogonally to the closest triangles to determine that which triangles "own" which points. Then we can calculate a density for each triangle like this:
Where
NOTE: If a single point corresponds to more than one triangle, we equally distribute it among the common triangles, so
is not necessarily a whole number.
Now we show an implementation of this method in Python. We use numpy arrays and a method from scipy, so they have to be imported:
import numpy as np
from scipy.spatial import KDTree
First we implement a function for calculating the areas of the triangles:
def triangle_areas(x, y, z, i, j, k):
v1x = x[j] - x[i]
v2x = x[k] - x[i]
v1y = y[j] - y[i]
v2y = y[k] - y[i]
v1z = z[j] - z[i]
v2z = z[k] - z[i]
crossx = v1y * v2z - v1z * v2y
crossy = v1z * v2x - v1x * v2z
crossz = v1x * v2y - v1y * v2x
areas = 0.5 * np.sqrt(crossx**2 + crossy**2 + crossz**2)
return areas
This returns a numpy with numbers corresponding to the areas of the triangles.
Then we implement the projector method in_triangle which determines if the given px, py, pz point projects to the triangle or not:
# Helper functions
def cross_prod(v1, v2):
return (v1[1] * v2[2] - v1[2] * v2[1], v1[2] * v2[0] - v1[0] * v2[2], v1[0] * v2[1] - v1[1] * v2[0])
def dot_prod(v1, v2):
return v1[0] * v2[0] + v1[1] * v2[1] + v1[2] * v2[2]
def in_triangle(x1, y1, z1, x2, y2, z2, x3, y3, z3, px, py, pz):
v1 = (x2 - x1, y2 - y1, z2 - z1)
v2 = (x3 - x2, y3 - y2, z3 - z2)
v3 = (x1 - x3, y1 - y3, z1 - z3)
normal = cross_prod(v1, v2)
in1 = cross_prod(v1, normal)
in2 = cross_prod(v2, normal)
in3 = cross_prod(v3, normal)
rel1 = ((-x1.T + px).T, (-y1.T + py).T, (-z1.T + pz).T)
rel2 = ((-x2.T + px).T, (-y2.T + py).T, (-z2.T + pz).T)
rel3 = ((-x3.T + px).T, (-y3.T + py).T, (-z3.T + pz).T)
tr1 = (dot_prod(rel1, in1) <= 0)
tr2 = (dot_prod(rel2, in2) <= 0)
tr3 = (dot_prod(rel3, in3) <= 0)
return np.logical_and(np.logical_and(np.equal(tr1, tr2), np.equal(tr1, tr3)), np.equal(tr2, tr3))
A function for smoothing the results is also built in for the sake of robustness:
def gauss_blur_point_set(x, y, z, values, sigma=1.0):
result_values = []
for i in range(len(x)):
d = ((x[i] - x)**2 + (y[i] - y)**2 + (z[i] - z)**2)
result_values.append(np.sum((np.e ** (-d / sigma)) * np.array(values)))
return np.array(result_values)
This just applies a filter on the values in space according to the Gaussian function.
Now we can implement the function that evaluates the surface density:
def points_on_face(x, y, z, i, j, k, points_x, points_y, points_z, blur=False, blur_sigma=1.0):
x = np.array(x)
y = np.array(y)
z = np.array(z)
i = np.array(i)
j = np.array(j)
k = np.array(k)
# Center of mass points
com_x = (x[i] + x[j] + x[k]) / 3.0
com_y = (y[i] + y[j] + y[k]) / 3.0
com_z = (z[i] + z[j] + z[k]) / 3.0
# Transform them to be able to use them in KDTree
com_points = np.array([com_x, com_y, com_z]).T
points = np.array([points_x, points_y, points_z]).T
tree = KDTree(com_points)
_, idx = tree.query(points, 7)
point_indices = np.fromiter(range(len(idx)), dtype="int")
is_eligible = in_triangle(x[i[idx]], y[i[idx]], z[i[idx]],
x[j[idx]], y[j[idx]], z[j[idx]],
x[k[idx]], y[k[idx]], z[k[idx]],
points_x[point_indices], points_y[point_indices], points_z[point_indices])
counts = np.zeros(len(i), dtype="float64")
for idx_toadd in range(len(idx)):
eligible_sum = np.sum(is_eligible[idx_toadd])
if eligible_sum != 0:
counts[idx[idx_toadd][is_eligible[idx_toadd]]
] += (1.0 / eligible_sum)
areas = triangle_areas(x, y, z, i, j, k)
# Normalize with the triangle areas
result = counts / areas
# If we want to apply the filter, then apply it wiht sigma smoothing
if blur:
result = gauss_blur_point_set(com_x, com_y, com_z, result, blur_sigma)
return result
We pass the mesh in the x, y, z, i, j, k arguments and the points from which the density is determined, are given in three arrays points_x, points_y, points_z. blur can be set to True to apply the filter.
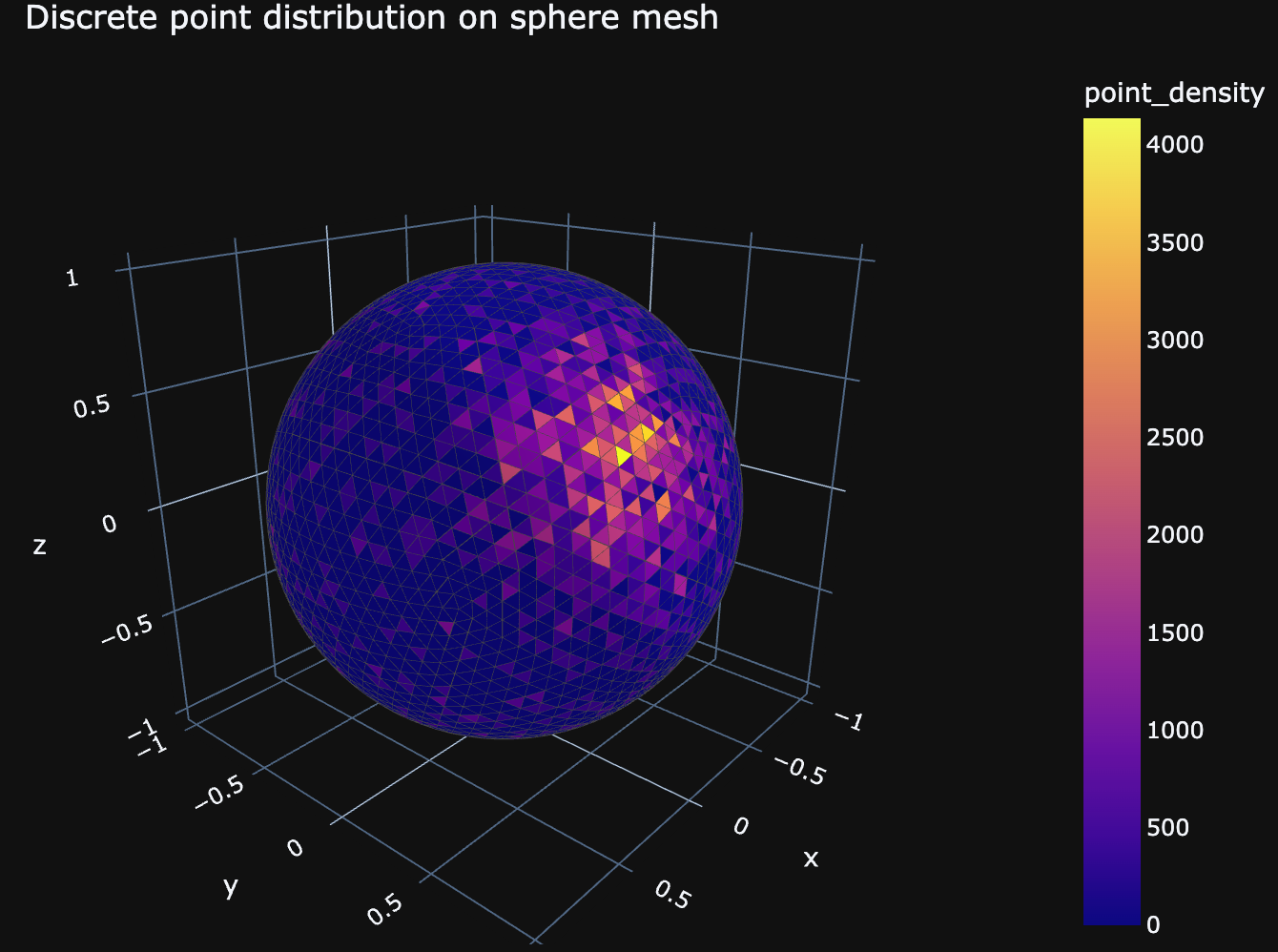
Let's say we generated a sphere from an icosahedron and we also have a function that generates random points isotropically on the surface of the sphere with a "bump" where we have a lot more points localized. This function looks like this:
import numpy.random as random
def generate_randoms_on_sphere_with_bump(seed=0, N = 1000, bump_N = 200, bump_phi = 0, bump_theta = np.pi/2., bump_size=0.1):
random.seed(seed)
points_z = (random.rand(N) - 0.5) * 2.0
thetas = np.arccos(points_z)
phis = random.rand(N) * (2 * np.pi)
points_x = np.cos(phis) * np.sin(thetas)
points_y = np.sin(phis) * np.sin(thetas)
bump_phis = random.normal(bump_phi, bump_size, bump_N)
bump_thetas = random.normal(bump_theta, bump_size, bump_N)
bump_z = np.cos(bump_thetas)
bump_y = np.sin(bump_phis) * np.sin(bump_thetas)
bump_x = np.cos(bump_phis) * np.sin(bump_thetas)
Where the arguments mean the following:
seed=0: the seed for random number generationN: number of points to generate isotropicallybump_N: number of points in the bumpbump_phi: the center bump_theta: the center bump_size: characterizes the size of the dumpThen we can apply all these together:
x, y, z, i, j, k = generate_sphere_fibonacci(1, 1500)
x, y, z, i, j, k = generate_icosahedron(1)
x, y, z, i, j, k = subdivide_triangles(x, y, z, i, j, k, 4)
for n in range(len(x)):
R = np.sqrt(x[n]**2 + y[n]**2 + z[n]**2)
x[n] = x[n]/R
y[n] = y[n]/R
z[n] = z[n]/R
points_x, points_y, points_z = generate_randoms_on_sphere_with_bump(2324, 1000, 900, bump_phi=1.2, bump_theta=1.1, bump_size=0.3)
counts = points_on_face(x, y, z, i, j, k, points_x, points_y, points_z, blur=False, blur_sigma=0.01)
And if we visualize the calculated densities, we get the following figure:

Which was done with the following code snippet:
import plotly.graph_objects as go
# Create wireframe lines
Xe = []
Ye = []
Ze = []
for face_n in range(len(i)):
Xe.append(x[i[face_n]])
Xe.append(x[j[face_n]])
Xe.append(x[k[face_n]])
Ye.append(y[i[face_n]])
Ye.append(y[j[face_n]])
Ye.append(y[k[face_n]])
Ze.append(z[i[face_n]])
Ze.append(z[j[face_n]])
Ze.append(z[k[face_n]])
Xe.append(None)
Ye.append(None)
Ze.append(None)
fig = go.Figure(data=[
go.Mesh3d(
x=x, y=y, z=z,
# i, j and k give the vertices of triangles
i=i, j=j, k=k,
name='y',
showscale=True,
colorbar_title='point_density',
# Intensity of each fac
intensity = counts,
intensitymode='cell',
),
go.Scatter3d(
x=Xe,
y=Ye,
z=Ze,
mode='lines',
name='',
line=dict(color= 'rgb(70,70,70)', width=1)
),
go.Scatter3d(
x=points_x,
y=points_y,
z=points_z,
mode='markers',
showlegend=False,
visible=False,
opacity=1,
),
])
fig.update_layout(title='Discrete point distribution on sphere mesh', margin={'autoexpand': True,'b': 5,'l': 5,'r': 5,'t': 30,}, autosize=True, width=700, height=500, template="plotly_dark")
fig.show()